Mohamed Aborageh
Mendelian Randomization for Estimating the Causal Impact of Comorbidities on Developing Parkinson's Dementia
Mohamed Aborageh shares a preview of his upcoming paper, in which he demonstrates how genetic data can be used to assess the causal effect that different comorbidities have on the development of dementia in Parkinson's patients.
Parkinson’s Dementia
Parkinson's Disease (PD) is a rapidly increasing neurological disorder and the second most prevalent neurodegenerative condition. It is mainly recognized by core motor symptoms such as bradykinesia, resting tremor, and rigidity, but cognitive deficits are also common among patients with PD (PwPD). Around 25% of PwPD experience mild cognitive impairment, and some develop PD dementia within a decade of diagnosis. PD dementia significantly raises healthcare costs and reduces quality of life, highlighting its importance to both patients and healthcare systems. Additionally, PD dementia is one of four critical milestones that typically appear about four years before death, indicating the onset of the terminal phase of the disease. Despite extensive research into the genetic underpinnings of PD, the reasons why some PwPD develop cognitive impairment early in the disease course remain unclear. Epidemiological studies report varying rates of dementia among PwPD, ranging from 20-80%, with different onset times. Genome-wide association studies (GWAS) have identified potential links between various genetic variants and cognitive impairment in PD, including variants in genes coding the proteins SNCA, MAPT, TMEM175, and GBA. However, GWAS may not always have enough power to detect rarer variants. Issues like linkage disequilibrium -where two different alleles are non-randomly associated and are detected together at the same loci with similar frequencies - and confounding effects - where external factors have effects on the outcome, and therefore distort the true effect of the exposure - can obscure the true impact of a variant on the phenotype.
Mendelian Randomization
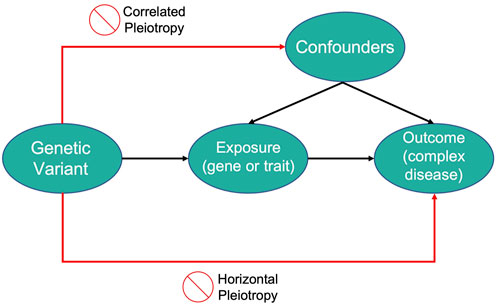
Mendelian Randomization (MR) is a technique that utilizes genetic variation to examine the causal effects of a modifiable exposure on disease or health outcomes. In MR studies, genetic variants serve as instrumental variables (IVs), defined as variables that affect an outcome solely through their strong association with an intermediary variable (Burgess et al., 2019, Sanderson et al., 2022).
The goal of MR studies is not to discover genetic variants directly linked to the disease but to use these variants as IVs for the modifiable exposure of interest. For genetic variants to qualify as IVs in MR studies, they must meet three criteria:
- The variant must be associated with the modifiable exposure.
- The variant must be independent of confounders that affect the relationship between the modifiable exposure and the outcome (no correlated pleiotropy).
- The variant must be independent of the outcome, given the modifiable exposure and the confounding factors (no horizontal pleiotropy).
Thus, genetic variants that account for variations in an exposure can be used to infer how changes in that exposure might impact the disease outcome of interest.
We obtained genetic variants for our exposures using the IEU-OpenGWAS API for different comorbidities including stroke, hypertension, type-2 diabetes, depression and excessive daytime sleeping, and conducted one-sample MR, measuring genetic variants, exposures, and outcomes within our individual-level data. For our exposures, we adjusted the SNP-exposure estimates to our phenotypes and generated SNP-outcome estimates through association testing, accounting for age and sex as covariates. The instrumental variables were selected via clumping to include SNPs that are strongly associated with our exposures and independent of each other and the causal effects of our exposures were estimated using the inverse-variance weighted (IVW) method with fixed effects. For sensitivity analysis, we employed MR-Egger and MR-PRESSO methods. The MR-Egger method checks for directional pleiotropy, determining if genetic factors have pleiotropic effects on the outcome that average out to non-zero. The MR-PRESSO method investigates horizontal pleiotropy in multi-instrument summary level MR through three steps: a global test to detect horizontal pleiotropy, correction for horizontal pleiotropy by removing outliers, and testing for significant differences before and after outlier removal.
The following article reviews the field of genetics in Parkinson’s Disease: https://doi.org/10.3389/fmmed.2022.933383
As soon as the upcoming paper on Mendelian Randomization has been published, it will be linked here as well.